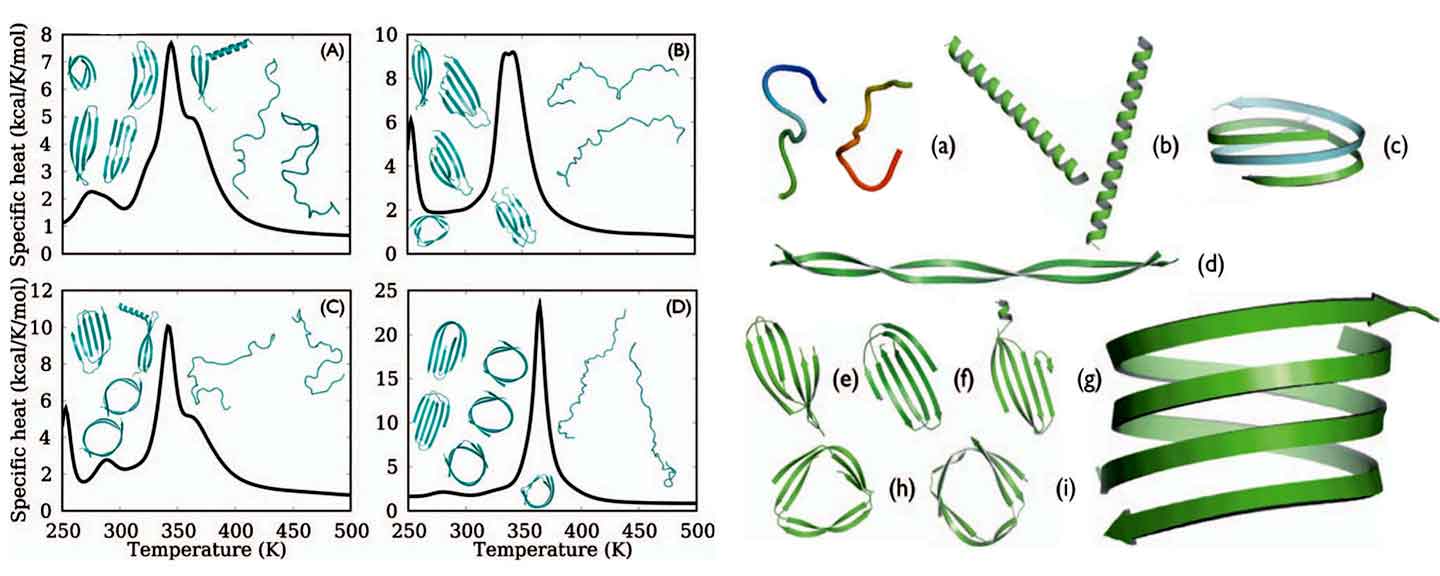
Applying the T-REMD simulation coupled with OPEP, we studied the aggregation process and structures of polyglutamine. Expansion of polyglutamine (polyQ) beyond the pathogenic threshold (35-40 Gln) is associated with several neurodegenerative diseases including Huntington’s disease. The polyglutamine monomer (single chain) and dimer with chain lengths from 30 to 50 residues have been studied starting from a random coil state. We obtained a complex picture for the first steps of aggregation that reproduces the
experimentally observed length-dependence of PQ and identifies the most likely amyloid building blocks for this sequence. The monomers does not show any significant β-stucture. For the dimer with 40 residues, by contrast, we observed spontaneous formation of antiparallel b-sheets and triangular and circular β-helical structures in a 50 temperatures ranging from 250-700 K and total integrated time 20 ms. The circular b-helical structure with ~32 Å diameter reorganizes further into a tight antiparallel double-stranded ~22 Å nanotube with 22 residues per turn and is very stable up to 350 K.
Spontaneous formation of polyglutamine nanotubes with molecular dynamics simulations
Rozita Laghaei and Normand Mousseau
J. Chem. Phys. 2010; 132, 165102
(This paper has been selected for the May 1, 2010 issue of Virtual Journal of Biological Physics Research and has also been selected for the May 10, 2010 issue of Virtual Journal of Nanoscale Science & Technology).