|
Tetrahedral
Finite Element Meshing for Bio-Molecules |
|
The following contains applications of our tetrahedral meshing scheme [1, 2] for mouse acetylcholinesterase (mAChE) and a tetramer AChE (mAChE_4) which consists of four mAChE subunits. |
|
|
|
1. mAChE |
|
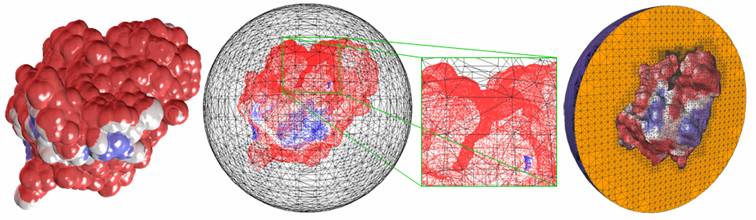
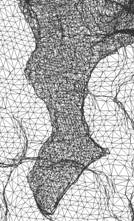
Figure 1: The molecular surface is extracted from
solvent accessibility data with an isovalue 0.5. The
color shows the distribution of the electrostatic potential over the
molecular surface (red is negative, blue positive, and white neutral). The
meshing is adaptive, and finer around the active gorge area. The solvent accessibility and electrostatic
potential data were generated by N. Baker et. al. |
|
2. mAChE_4 |
|
|
|
|
|
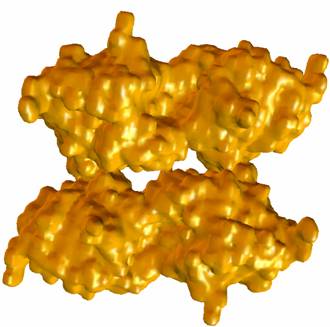
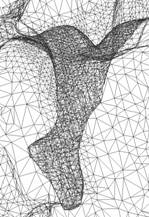
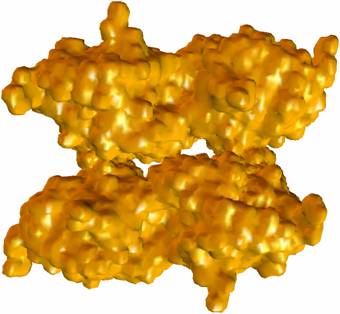
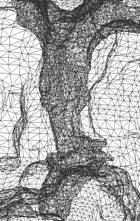
Figure 2: Adaptive finite element tetrahedral meshes for a mAChE_4 molecular surface. The molecular surface is extracted from a solvent accessibility map (257^3), of C. Bajaj et.al, with an isovalue of 1.0. |
|
|
|
|
|
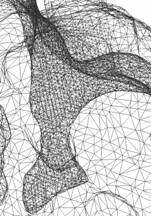
Figure 3: Adaptive finite element mesh of a mAChE_4 molecular surface. The molecular surface is extracted from the solvent accessibility map of Nathan Baker, David Zhang et al (449^3), with an isovalue 0.5. |
|
|
|
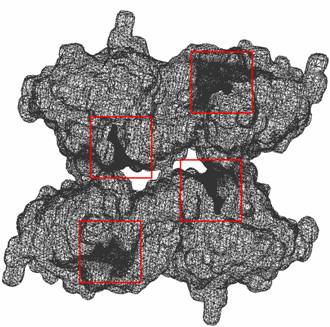
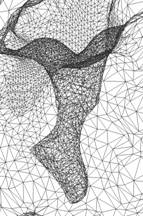
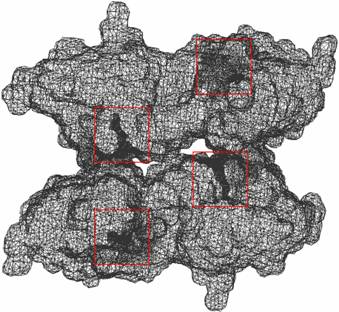
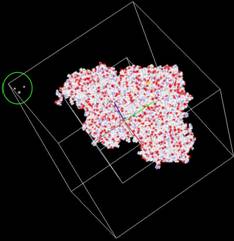
3. Problems: (1) We found three small components at the Left_Top corner, which are perhaps spurious hetatms. The pqr file could be corrected. |
|
Figure 4: The visualizations are generated from our Volume Rover and our LBIE Mesh generation software. |
|
|
|
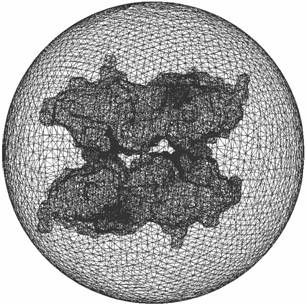
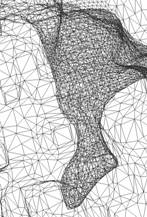
(2) For the current mesh, our chosen bounding sphere for a 513^3 accessibility map, part of the molecular surface is outside the bounding sphere. We shall enlarge the bounding sphere for the exterior tetrahedral mesh, or regenerate a little smaller dataset.
Figure 5: The molecular surface and an outer sphere
within 513^3 cube. |
|
|
|
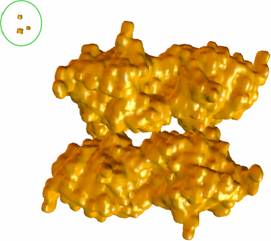
(3) We also tried half-resolution data of 449^3 solvent accessibility data. The four gorges of mAChE_4 can be reconstructed and adaptively meshed, with almost the same topology (minor local modification is done in postprocessing the mesh, and similar to what we did for mAChE and papers [3, 4]). |
|
|
|
References: 1. Y. Zhang, C. Bajaj, B-S. Sohn. 3D Finite Element Meshing from Imaging Data. Submitted to the special issue of Computer Methods in Applied Mechanics and Engineering (CMAME) on Unstructured Mesh Generation, 2003. 2. Y.
Zhang, C. Bajaj, B-S. Sohn.
Adaptive and Quality 3D Meshing from Imaging Data. Proceedings of
8th ACM Symposium on Solid Modeling and Applications, pp. 286-291. 3. Y. Song, Y. Zhang, C. Bajaj, N. Baker. Continuum Diffusion Reaction Rate Calculations of Wild Type and Mutant Mouse Acetylcholinesterase: Adaptive Finite Element Analysis. Submitted to Biophysical Journal, 2004. 4. Y. Song, Y. Zhang, T. Shen, C. Bajaj, J. McCammon, N. Baker. Finite Element Solution of the Steady-state Smoluchowski Equation for Rate Constant Calculations. Biophysical Journal, Vol. 86, No. 4, pp. 1-13, April 2004. |
|
*
Questions about these images, should be directed to jessica@ices.utexas.edu. |